Single Cell mRNA Seq Work in Leukaemia
Fri, 22nd November 2024 | 8PM - 9:30PM (GMT+8 SG Time)*
Join our expert panel from the Wellcome Sanger Institute and University of Cambridge, UK, as they share groundbreaking research in single-cell mRNA sequencing and its applications in leukaemia.
Discover the latest findings from renowned experts Prof Sam Behjati, who will discuss the importance of single leukaemia cell transcriptomes, and Dr Laura Jardine, who will present on normal reference atlases from human foetuses. Additionally, Mr Bram Lim (A*STAR funded MB/PhD student) and Ms Mi Trinh (PhD student) will showcase cutting-edge research in childhood T-AL and myeloid leukaemia of Down’s syndrome.
Secure your spot now and gain valuable insights into the future of leukaemia research!
PRESENTATION SUMMARIES
Why study single leukaemia cell transcriptomes? by Prof Sam Behjati
A decade of “omic” research into childhood leukaemia has provided answers to many fundamental questions of leukaemia research. However, there remain significant gaps in our knowledge that coalesce around the central question of the leukaemia cell actually is. The advent of high throughput single cell genomics has enabled us to study leukaemia cells in an unbiased way and at unprecedented resolution. In this lecture I will provide an introduction into single leukaemia cell mRNA sequencing.
Normal reference atlases from human foetuses by Dr Laura Jardine
This presentation will cover our insights from single cell multiomic studies of human blood haematopoiesis with respect to B acute lymphoblastic leukaemia and transient abnormal myelopoiesis of Down syndrome.
A non-canonical lymphoblast in refractory childhood T-ALL by Mr Bram Lim
Childhood T-cell acute lymphoblastic leukaemia (T-ALL) is an aggressive malignancy which carries a worse clinical prognosis than its B-cell counterpart (B-ALL). Despite extensive genomic characterisation, no robust genetic or phenotypic marker has been identified for accurate clinical risk stratification of T-ALL at the point of diagnosis, unlike in B-ALL. Here, we performed single-cell RNA sequencing (scRNA-seq) of T-ALL from children who did or did not respond to initial treatment. We identified a transcriptionally distinctive blast population, exhibiting features of innate-like lymphocytes, as the major source of refractory disease. Evidence of such blasts at diagnosis heralded refractory disease across independent datasets and was associated with survival in a large, contemporary trial cohort. Moreover, for one individual, by projecting somatic mutations onto scRNA-seq data, we demonstrated that these non-canonical blasts were detectable at diagnosis as a minor subclone (~1% of blasts), but expanded dramatically following treatment to become the dominant clone. Our findings portray refractory T-ALL as a distinct disease driven by a previously unknown lymphoblast variant and may have immediate clinical utility.
Example myeloid leukaemia of Down’s syndrome by Ms Mi Trinh
Children born with Down syndrome, a genetic condition caused by the presence of an additional chromosome 21 (trisomy 21 or T21), have a substantially higher risk of developing childhood leukaemia. Approximately 15-30% of infants with Down syndrome develop a preleukemic condition called transient abnormal myelopoiesis (TAM). TAM is strictly associated with GATA1 truncating mutations on a T21 background. Most TAM cases spontaneously resolve within the first few months of life. However, by the age of five, approximately 10% of cases progress to megakaryoblastic/erythroid leukaemia, known as myeloid leukaemia of Down syndrome (ML-DS). This final step of transformation is often driven by specific “third-hit” somatic mutations, most of which affects genes encoding JAK kinases, cohesin complexes, or epigenetic regulators. These well-defined stepwise genetic aberrations associated with ML-DS pathogenesis provide a unique and highly tractable system to elucidate the molecular changes underlying leukaemogenesis.
Using single-cell mRNA sequencing of primary human samples, we directly interrogate the transcriptional changes underpinning the multi-step pathogenesis of ML-DS. We observe an expansion of megakaryocyte-erythrocyte progenitors (MEPs) specifically in T21 foetal liver compared to diploid or other trisomies. However, it is the GATA1 mutations that accounts for the majority of the transcriptomic changes towards that of the leukaemic blasts. Furthermore, investigation of eventful TAM cases reveals an enrichment in expression of the leukaemogenic gene module which is widely utilised in ML-DS and other leukaemia. Finally, by integrating the genomic and transcriptomic data, we directly defined the genomic evolution, and its corresponding transcriptional consequences, that underpins a treatment-refractory case of ML-DS. Overall, we have generated the first comprehensive single-cell mRNA atlas of TAM/ML-DS, enabling unbiased quantitative characterisation of the transcriptional evolution along ML-DS pathogenesis, as well as transcriptional features of progressive TAM. These insights offer the possibility of more meaningful clinical interpretations and the potential to explore further clinical hypotheses through large-scale international studies.
WATCH IT AGAIN
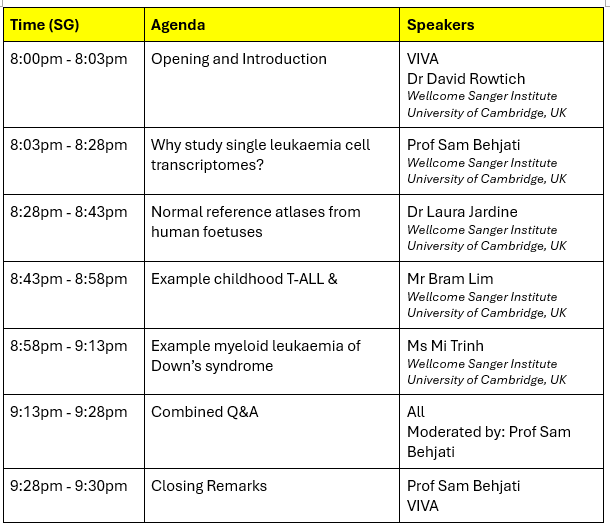
PROGRAMME RUNDOWN
SPEAKERS

Prof Sam Behjati
Honorary consultant paediatric oncologist
Clinical Professor of Paediatric Oncology,
University of Cambridge Group Leader, Wellcome Sanger Institute
United Kingdom

Dr Laura Jardine
Associate Clinical Lecturer
Newcastle University
Wellcome Sanger Institute,
University of Cambrdige
United Kingdom

Mr Bram Lim
PhD Student,
Wellcome Sanger Institute,
University of Cambrdige
United Kingdom

Ms Mi Trinh
PhD Student,
Wellcome Sanger Institute,
University of Cambrdige
United Kingdom
MODERATOR

Prof Sam Behjati
Honorary consultant paediatric oncologist
Clinical Professor of Paediatric Oncology,
University of Cambridge Group Leader, Wellcome Sanger Institute
United Kingdom